Blinded with Science or Informed by Charts? Annotated Analysis Scripts of a Replication Study
Pierre Dragicevic and Yvonne Jansen
All scripts were first generated in March 2017, then cleaned up and marked up in August/September 2017
In this document we detail the planned and additional analyses for our experiments. By default, all code elements are hidden and only the R output is visible. Individual code segments can be made visible through the respective buttons. To rerun complete analyses we recommend to download our zip file containg all analysis scripts.
Experiment 1
# Clean up memory
rm(list = ls())
# Experiment filenames
results_file <- "exp1/results"
crowflower_file <- "exp1/crowdflower_data"
invalid_answers_file <- "exp1/invalid_answers"
figure_path <- "plots/exp1-"
# Stimuli values (used to compute comprehension errors)
control <- 87 # % sick people without the drug
drug <- 47 # % sick people with the drug
# compute the true answer given the data
true_answer <- drug/control * 20
# Values reported by Tal and Wanksink to add to our plots as a comparison
bws_nochart <- 6.12
bws_chart <- 6.83
bws_p <- 0.04Planned Analysis
library("plyr")
library("dplyr")##
## Attaching package: 'dplyr'## The following objects are masked from 'package:plyr':
##
## arrange, count, desc, failwith, id, mutate, rename, summarise,
## summarize## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, unionsource("CI.helpers.R")## Simple Bootstrap Routines (1.1-3 2008-04-30)source("misc.helpers.R")
source("plotting functions.R")##
## Attaching package: 'gridExtra'## The following object is masked from 'package:dplyr':
##
## combine##
## Attaching package: 'cowplot'## The following object is masked from 'package:ggplot2':
##
## ggsave# indicated whether we want ggplot to save the generated plots
save_plots <- FALSE
# print plots to screen
print_plots <- TRUELoad data
# Load experiment data from csv file
if (exists("results_file")) {
data <- read.csv(paste("data/", results_file, ".csv", sep = ""), sep = ",")
}
# Add crowdflower metadata (if exists)
if (exists("crowflower_file")) {
crowdsourcedata <- read.csv(paste("data/", crowflower_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$completion_code <- as.character(crowdsourcedata$completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "completion_code"))
# Load country codes
country_file <- "countries"
countrydata <- read.csv(paste("data/", country_file, ".csv", sep = ""),
sep = ",")
}
# Add mturk metadata (if exists)
if (exists("mturk_file")) {
crowdsourcedata <- read.csv(paste("data/", mturk_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$Answer.completion_code <- as.character(crowdsourcedata$Answer.completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "Answer.completion_code"))
}Invalid jobs
# Utility function for printing percentages
percent <- function(x, n) {
if (x/n < 0.01)
r <- 1 else r <- 0
paste(" (", format_number(100 * x/n, r), "%)", sep = "")
}
print.title("Invalid jobs")##
## ###################################
## # Invalid jobs
## ###################################data_invalid <- read.csv(paste("data/", invalid_answers_file, ".csv", sep = ""),
sep = ",")
n_valid <- nrow(data)
n_invalid <- nrow(data_invalid)
n <- n_valid + n_invalid
cat("Valid jobs: ", n_valid, "\n")## Valid jobs: 123cat("Invalid jobs: ", n_invalid, "\n\n")## Invalid jobs: 25#
n_failed_attention <- nrow(data_invalid[data_invalid$test_question != "common_cold",
])
n_abnormal_time <- nrow(data_invalid[data_invalid$time_taken < 30 | data_invalid$time_taken >
15 * 60, ])
n_previous_study <- nrow(data_invalid[data_invalid$previous_study == "yes",
])
cat("Failed attention check: ", n_failed_attention, percent(n_failed_attention,
n), "\n", sep = "")## Failed attention check: 7 (5%)cat("Abnormal completion time: ", n_abnormal_time, percent(n_abnormal_time,
n), "\n", sep = "")## Abnormal completion time: 4 (3%)cat("Did study before: ", n_previous_study, percent(n_previous_study,
n), "\n", sep = "")## Did study before: 18 (10%)Sample size, completion time and demographics
# Sample size
N <- nrow(data)
cat("Total sample size: ", N, "\n")## Total sample size: 123cat("n =", sum(data$condition == "no_graph"), "for the no-chart condition\n")## n = 62 for the no-chart conditioncat("n =", sum(data$condition == "graph"), "for the chart condition\n\n")## n = 61 for the chart condition# Job completion time
mt <- median(data$time_taken)
cat("Median job completion time:", format_number(mt/60, 2), "minutes.\n\n")## Median job completion time: 2.4 minutes.# Countries
if (exists("crowflower_file")) {
data$country <- sapply(data$X_country, function(code) countrydata$name[match(code,
countrydata$alpha.3)])
data$subregion <- sapply(data$X_country, function(code) countrydata$sub.region[match(code,
countrydata$alpha.3)])
data$continent <- sapply(data$X_country, function(code) countrydata$region[match(code,
countrydata$alpha.3)])
cat("Number of different countries:", length(unique(data$country)), "\n")
print.percents(data$continent)
}## Number of different countries: 32
## n %
## 1 Europe 68 55
## 2 Americas 34 28
## 3 Asia 21 17# Gender distribution
# Column is present in all but experiment 3
if ("gender" %in% colnames(data)) {
cat("\nGender:\n")
print.percents(data$gender)
}##
## Gender:
## n %
## 1 male 80 65
## 2 female 43 35# Age
# Column is present in all but experiment 3
if ("age" %in% colnames(data)) {
cat("\nMean age:", round(mean(data$age)), "\n")
}##
## Mean age: 35# Education
# Column is present in all but experiment 3
if ("education" %in% colnames(data)) {
cat("\nEducation:\n")
print.percents(data$education)
}##
## Education:
## n %
## 1 college_or_more 59 48
## 2 some_college 34 28
## 3 high-school 27 22
## 4 neither 3 2Analysis of dependent variables
##### Retrieve data
# Extract judgments of drug efficacy (values between 1 and 9). Experiment 1
# and experiments 2-4 use different question wordings, hence we look at two
# possible columns.
# Column is present in experiment 1, which replicates the question from BwS
# experiment 1
if ("effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$effectiveness
eff_graph <- data[data$condition == "graph", ]$effectiveness
variable_name <- "Perceived effectiveness"
}
# Column is present in experiments 2-4, which replicate the question from
# BwS experiment 2
if ("believe_in_effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$believe_in_effectiveness
eff_graph <- data[data$condition == "graph", ]$believe_in_effectiveness
variable_name <- "Belief in efficacy"
}
# Extract belief in drug from the second question (yes = not effective, no =
# effective).
# Column is present in experiment 1 only.
if ("reduces_sickness" %in% colnames(data)) {
yesno_to_01 <- function(x) {
if (x == "yes")
1 else if (x == "no")
0 else NA
}
ill_nograph <- data[data$condition == "no_graph", ]$reduces_sickness
ill_graph <- data[data$condition == "graph", ]$reduces_sickness
ill_nograph <- sapply(ill_nograph, yesno_to_01)
ill_graph <- sapply(ill_graph, yesno_to_01)
}
# Extract answers to comprehension question
num_nograph <- data[data$condition == "no_graph", ]$comprehension
num_graph <- data[data$condition == "graph", ]$comprehension
##### Descriptive and inferential statisticsJudgment of efficacy
##### CIs for judgment of efficacy on a 1--9 scale
mean_eff_nograph <- meanCI.bootstrap(eff_nograph)
mean_eff_graph <- meanCI.bootstrap(eff_graph)
mean_eff_diff <- diffMeanCI.bootstrap(eff_graph, eff_nograph)
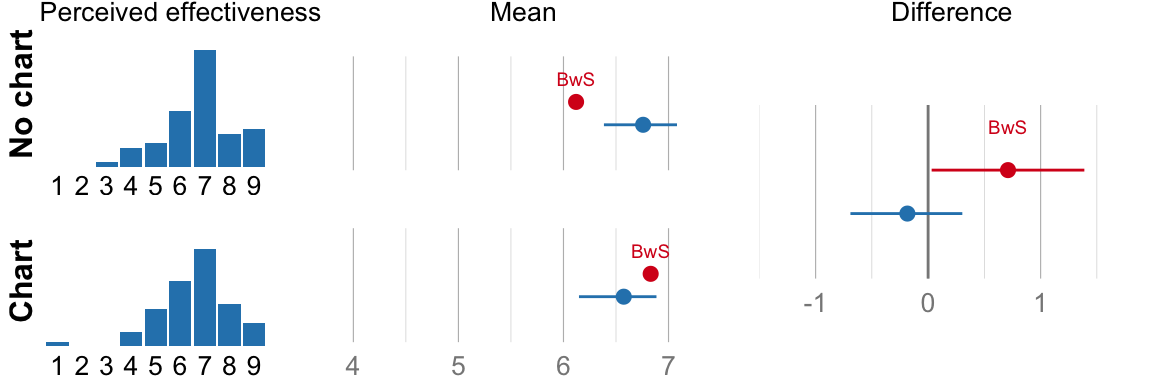
print.title(variable_name)##
## ###################################
## # Perceived effectiveness
## ###################################cat("Mean efficacy no-chart: ", format_ci(mean_eff_nograph), "\n")## Mean efficacy no-chart: 6.8, CI [6.4, 7.1]cat("Mean efficacy chart: ", format_ci(mean_eff_graph), "\n")## Mean efficacy chart: 6.6, CI [6.1, 6.9]cat("Difference: ", format_ci(mean_eff_diff), "\n")## Difference: -0.18, CI [-0.69, 0.30]# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
ttest <- t.test(eff_graph, eff_nograph)
mean_eff_diff_normal <- c(mean(eff_graph) - mean(eff_nograph), ttest$conf.int[1],
ttest$conf.int[2])
cat("(using a t-test) ", format_ci(mean_eff_diff_normal), "p =", format_number(ttest$p.value),
"\n")## (using a t-test) -0.18, CI [-0.69, 0.33] p = 0.48# Display distributions of efficacy judgments and confidence intervals for
# estimates
## compile CIs and BwS results into a dataframe for plotting
eff_df <- NULL
eff_df <- add.ci.to.df(mean_eff_nograph, "no chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_nochart, bws_nochart, bws_nochart), "no chart",
"BwS", eff_df)
eff_df <- add.ci.to.df(mean_eff_graph, "chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_chart, bws_chart, bws_chart), "chart", "BwS", eff_df)
eff_diff_df <- NULL
eff_diff_df <- add.ci.to.df(mean_eff_diff, " ", "ours")
eff_diff_df <- add.ci.to.df(ci_from_p(bws_chart - bws_nochart, bws_p), " ",
"BwS", eff_diff_df)
if (variable_name == "Belief in efficacy") {
# plotting for experiments 2-4
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp24,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "believe_in_efficacy_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
} else {
# plotting for experiment 1
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.1.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp1,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "perceived_effectiveness_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
}
Binary belief in efficacy
##### CIs for binary belief in efficacy
# Column is present in experiment 1 only, as subsequent experiments didn't
# include that question.
if ("reduces_sickness" %in% colnames(data)) {
mean_ill_nograph <- propCI(sum(ill_nograph == 1), length(ill_nograph))
mean_ill_graph <- propCI(sum(ill_graph == 1), length(ill_graph))
mean_ill_diff <- diffpropCI(sum(ill_graph == 1), length(ill_graph), sum(ill_nograph ==
1), length(ill_nograph))
print.title("Belief in efficacy (binary)")
cat("Proportion of 'yes' no-chart: ", format_ci(mean_ill_nograph, percent = TRUE),
"\n")
cat("Proportion of 'yes' chart: ", format_ci(mean_ill_graph, percent = TRUE),
"\n")
cat("Difference: ", format_ci(mean_ill_diff, percent = TRUE),
"\n")
# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
yes <- c(sum(ill_graph == 1), sum(ill_nograph == 1))
all <- c(length(ill_graph), length(ill_nograph))
proptest <- prop.test(yes, all, correct = F) # disable continuity correction to get the same p-value
mean_ill_diff_2 <- c(yes[1]/all[1] - yes[2]/all[2], proptest$conf.int[1],
proptest$conf.int[2])
cat("(using an X-squared test) ", format_ci(mean_ill_diff_2, percent = TRUE),
"p =", format_number(proptest$p.value), "\n")
# Plot our results in comparison to the BwS results
# Tal and Wanksink's results
bws.cis.no_graph <- propCI(21, 31)
bws.cis.graph <- propCI(28, 29)
bws.cis.diff <- diffpropCI(28, 29, 21, 31)
# construct the dataframes for plotting
efficiency.df <- NULL
efficiency.df <- add.ci.to.df(mean_ill_nograph, "no chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(mean_ill_graph, "chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.no_graph, "no chart", "BwS", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.graph, "chart", "BwS", efficiency.df)
efficiency.diff.df <- NULL
efficiency.diff.df <- add.ci.to.df(mean_ill_diff, " ", "ours", efficiency.diff.df)
efficiency.diff.df <- add.ci.to.df(bws.cis.diff, " ", "BwS", efficiency.diff.df)
# plot
efficiency.ciplot <- efficacy.belief.ciplot(efficiency.df, efficiency.diff.df)
efficiency.plot <- drawHorizontalEfficiency(efficiency.ciplot, "Belief in efficacy",
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "belief_in_efficacy_plot.pdf"),
efficiency.plot, base_aspect_ratio = 4, base_height = 1.5)
if (print_plots)
print(efficiency.plot)
}##
## ###################################
## # Belief in efficacy (binary)
## ###################################
##
## Proportion of 'yes' no-chart: 90%, CI [80%, 95%]
## Proportion of 'yes' chart: 87%, CI [76%, 93%]
## Difference: -3.4%, CI [-16%, 8.4%]
## (using an X-squared test) -3.4%, CI [-15%, 7.8%] p = 0.55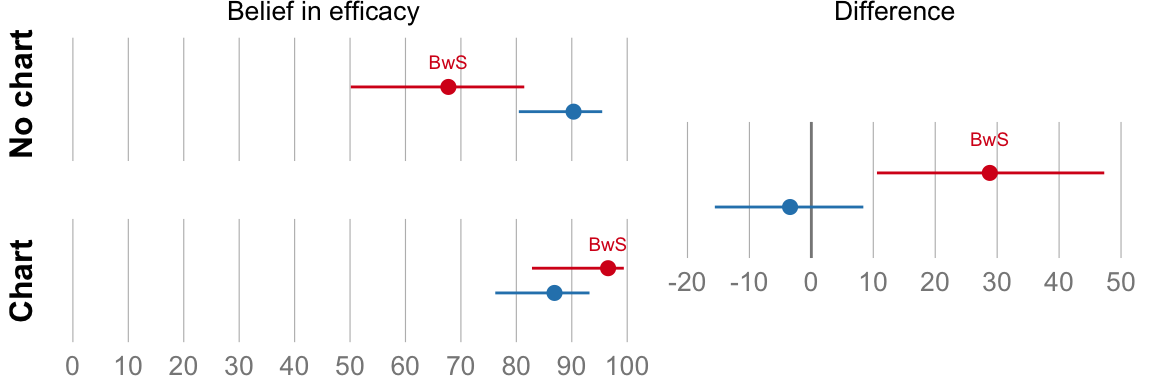
Comprehension question
#### CIs for errors at the comprehension question
# Define error
psick <- drug/control
logodds <- function(p) {
log(p/(1 - p))
}
logodds.psick <- logodds(psick)
error <- function(n) {
n[n == 0] <- 0.1
n[n == 20] <- 19.9
abs(logodds(n/20) - logodds.psick)
}
# Compute errors
data$error <- error(data$comprehension)
err_nograph <- error(num_nograph)
err_graph <- error(num_graph)
# Use geometric mean to reduce the influence of outliers and increase the
# weight of close-to-exact answers
mean_err_nograph <- geomMeanCI.bootstrap(err_nograph)
mean_err_graph <- geomMeanCI.bootstrap(err_graph)
mean_err_diff <- ratioGeomMeanCI.bootstrap(err_graph, err_nograph)
print.title("Comprehension error")##
## ###################################
## # Comprehension error
## ###################################cat("Geometric mean error no-chart: ", format_ci(mean_err_nograph), "\n")## Geometric mean error no-chart: 0.66, CI [0.49, 0.88]cat("Geometric mean error chart: ", format_ci(mean_err_graph), "\n")## Geometric mean error chart: 0.43, CI [0.33, 0.56]cat("Ratio: ", format_ci(mean_err_diff), "\n")## Ratio: 0.65, CI [0.44, 0.98]# Plot raw responses and confidence intervals
comp.hist.no_chart <- histo(num_nograph, 20, "no chart", true_answer, 1)
comp.hist.chart <- histo(num_graph, 20, "chart", true_answer, 1)
# construct the dataframes for plotting
comprehension.df <- NULL
comprehension.df <- add.ci.to.df(mean_err_nograph, "no chart", "ours", comprehension.df)
comprehension.df <- add.ci.to.df(mean_err_graph, "chart", "ours", comprehension.df)
comprehension.diff.df <- NULL
comprehension.diff.df <- add.ci.to.df(mean_err_diff, " ", "ours", comprehension.diff.df)
# plot
comp.cis <- comprehension.ciplot(comprehension.df, comprehension.diff.df)
comp.plot <- drawVerticalComp(comp.hist.no_chart, comp.hist.chart, comp.cis,
"Comprehension error", "Error ratio")
if (save_plots) save_plot(paste(sep = "", figure_path, "comprehension_plot.pdf"),
comp.plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots) print(comp.plot)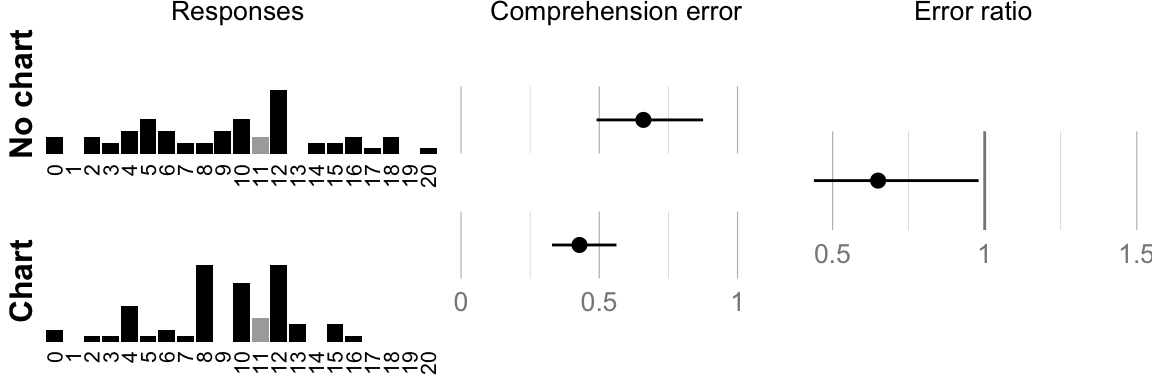
Additional Analysis
Bias in responses
cat("Mean response no-chart: ", format_ci(meanCI.bootstrap(num_nograph), sigdigits = 3),
"\n")## Mean response no-chart: 9.19, CI [7.91, 10.4]cat("Mean response chart: ", format_ci(meanCI.bootstrap(num_graph), sigdigits = 3),
"\n")## Mean response chart: 9.13, CI [8.14, 9.97]cat("Mean difference: ", format_ci(diffMeanCI.bootstrap(num_graph, num_nograph)),
"\n")## Mean difference: -0.062, CI [-1.67, 1.40]# Justifications from those who responded 'no' to the belief question. This
# data was not reported, but it informed the design of our polarization
# question (see section 6.1)Justifications to ‘no’ answers
whyno <- as.character(data$why_answered_no)
whyno <- whyno[whyno != ""]
cat(whyno, sep = "\n-> ")## this is how i think
## -> it depend on human body and immune system
## -> because more side effect of medication
## -> Not all people react the same to the medication. It takes a lot of time to demonstrate the usefulness of the medication.
## -> I do not belive that the medications can actually increase immunity. Drugs act on the symptoms.
## -> the drug busts the immune system. There is no way to know if this is the case nor if the user would get sick without it.
## -> i dont belive in miraculous drugs if they found it the studies where over by now
## -> I didn't compute it in the way that the follow up question did, which assumed that those 20 people would by default all get sick. I thought this lower sickness rate could simply be due to extraneous factors. And since the remaining percentage of people getting ill was still rather high, I chose NO here.
## -> It boost immune system still the illnes remains
## -> I simply don't believe in such medication, never had any proof.
## -> The medication does not reduce the disease
## -> The drug is implicitly stated as boosting the human immune system, not fight infections.
## -> Because you can't cure cold.
## -> Because it increases immunity, but does not cure diseaseExperiment 2
# Clean up memory
rm(list = ls())
# Experiment filenames
results_file <- "exp2/results"
crowflower_file <- "exp2/crowdflower_data"
invalid_answers_file <- "exp2/invalid_answers"
figure_path <- "plots/exp2-"
# Stimuli values (used to compute comprehension errors)
control <- 83 # % sick people without the drug
drug <- 63 # % sick people with the drug
# compute the true answer given the data
true_answer <- 63/83 * 20
# Values reported by Tal and Wanksink to add to our plots as a comparison
bws_nochart <- 4.66
bws_chart <- 5.75
bws_p <- 0.0066Planned Analysis
library("plyr")
library("dplyr")
source("CI.helpers.R")
source("misc.helpers.R")
source("plotting functions.R")
# indicated whether we want ggplot to save the generated plots
save_plots <- FALSE
# print plots to screen
print_plots <- TRUELoad data
# Load experiment data from csv file
if (exists("results_file")) {
data <- read.csv(paste("data/", results_file, ".csv", sep = ""), sep = ",")
}
# Add crowdflower metadata (if exists)
if (exists("crowflower_file")) {
crowdsourcedata <- read.csv(paste("data/", crowflower_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$completion_code <- as.character(crowdsourcedata$completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "completion_code"))
# Load country codes
country_file <- "countries"
countrydata <- read.csv(paste("data/", country_file, ".csv", sep = ""),
sep = ",")
}
# Add mturk metadata (if exists)
if (exists("mturk_file")) {
crowdsourcedata <- read.csv(paste("data/", mturk_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$Answer.completion_code <- as.character(crowdsourcedata$Answer.completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "Answer.completion_code"))
}Invalid Jobs
# Utility function for printing percentages
percent <- function(x, n) {
if (x/n < 0.01)
r <- 1 else r <- 0
paste(" (", format_number(100 * x/n, r), "%)", sep = "")
}
data_invalid <- read.csv(paste("data/", invalid_answers_file, ".csv", sep = ""),
sep = ",")
n_valid <- nrow(data)
n_invalid <- nrow(data_invalid)
n <- n_valid + n_invalid
cat("Valid jobs: ", n_valid, "\n")## Valid jobs: 164cat("Invalid jobs: ", n_invalid, "\n\n")## Invalid jobs: 32#
n_failed_attention <- nrow(data_invalid[data_invalid$test_question != "common_cold",
])
n_abnormal_time <- nrow(data_invalid[data_invalid$time_taken < 30 | data_invalid$time_taken >
15 * 60, ])
n_previous_study <- nrow(data_invalid[data_invalid$previous_study == "yes",
])
cat("Failed attention check: ", n_failed_attention, percent(n_failed_attention,
n), "\n", sep = "")## Failed attention check: 6 (3%)cat("Abnormal completion time: ", n_abnormal_time, percent(n_abnormal_time,
n), "\n", sep = "")## Abnormal completion time: 3 (2%)cat("Did study before: ", n_previous_study, percent(n_previous_study,
n), "\n", sep = "")## Did study before: 20 (10%)Sample size, completion time and demographics
# Sample size
N <- nrow(data)
cat("Total sample size: ", N, "\n")## Total sample size: 164cat("n =", sum(data$condition == "no_graph"), "for the no-chart condition\n")## n = 79 for the no-chart conditioncat("n =", sum(data$condition == "graph"), "for the chart condition\n\n")## n = 85 for the chart condition# Job completion time
mt <- median(data$time_taken)
cat("Median job completion time:", format_number(mt/60, 2), "minutes.\n\n")## Median job completion time: 2.0 minutes.# Countries
if (exists("crowflower_file")) {
data$country <- sapply(data$X_country, function(code) countrydata$name[match(code,
countrydata$alpha.3)])
data$subregion <- sapply(data$X_country, function(code) countrydata$sub.region[match(code,
countrydata$alpha.3)])
data$continent <- sapply(data$X_country, function(code) countrydata$region[match(code,
countrydata$alpha.3)])
cat("Number of different countries:", length(unique(data$country)), "\n")
print.percents(data$continent)
}## Number of different countries: 34
## n %
## 1 Europe 91 55
## 2 Americas 37 23
## 3 Asia 32 20
## 4 Africa 2 1
## 5 Oceania 2 1# Gender distribution
# Column is present in all but experiment 3
if ("gender" %in% colnames(data)) {
cat("\nGender:\n")
print.percents(data$gender)
}##
## Gender:
## n %
## 1 male 118 72
## 2 female 46 28# Age
# Column is present in all but experiment 3
if ("age" %in% colnames(data)) {
cat("\nMean age:", round(mean(data$age)), "\n")
}##
## Mean age: 34# Education
# Column is present in all but experiment 3
if ("education" %in% colnames(data)) {
cat("\nEducation:\n")
print.percents(data$education)
}##
## Education:
## n %
## 1 college_or_more 94 57
## 2 some_college 44 27
## 3 high-school 22 13
## 4 neither 4 2Analysis of dependent variables
##### Retrieve data
# Extract judgments of drug efficacy (values between 1 and 9). Experiment 1
# and experiments 2-4 use different question wordings, hence we look at two
# possible columns.
# Column is present in experiment 1, which replicates the question from BwS
# experiment 1
if ("effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$effectiveness
eff_graph <- data[data$condition == "graph", ]$effectiveness
variable_name <- "Perceived effectiveness"
}
# Column is present in experiments 2-4, which replicate the question from
# BwS experiment 2
if ("believe_in_effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$believe_in_effectiveness
eff_graph <- data[data$condition == "graph", ]$believe_in_effectiveness
variable_name <- "Belief in efficacy"
}
# Extract belief in drug from the second question (yes = not effective, no =
# effective).
# Column is present in experiment 1 only.
if ("reduces_sickness" %in% colnames(data)) {
yesno_to_01 <- function(x) {
if (x == "yes")
1 else if (x == "no")
0 else NA
}
ill_nograph <- data[data$condition == "no_graph", ]$reduces_sickness
ill_graph <- data[data$condition == "graph", ]$reduces_sickness
ill_nograph <- sapply(ill_nograph, yesno_to_01)
ill_graph <- sapply(ill_graph, yesno_to_01)
}
# Extract answers to comprehension question
num_nograph <- data[data$condition == "no_graph", ]$comprehension
num_graph <- data[data$condition == "graph", ]$comprehensionBelief in efficacy
##### CIs for judgment of efficacy on a 1--9 scale
mean_eff_nograph <- meanCI.bootstrap(eff_nograph)
mean_eff_graph <- meanCI.bootstrap(eff_graph)
mean_eff_diff <- diffMeanCI.bootstrap(eff_graph, eff_nograph)
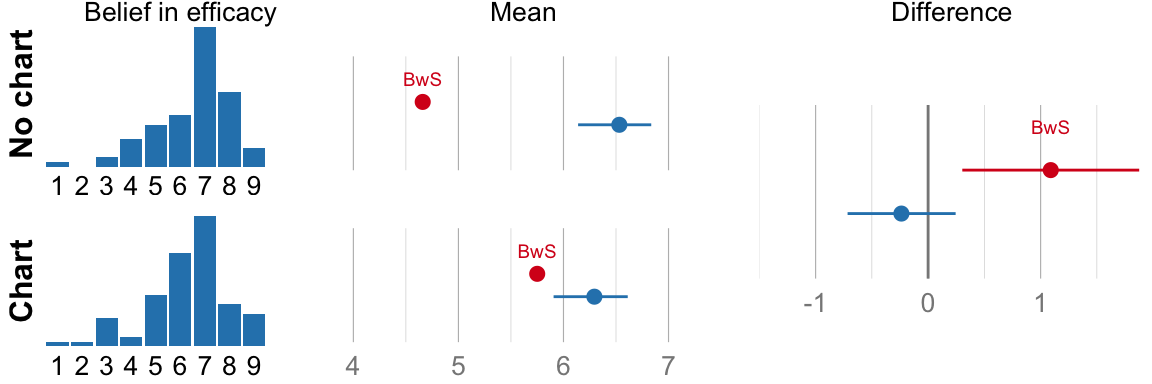
cat("Mean efficacy no-chart: ", format_ci(mean_eff_nograph), "\n")## Mean efficacy no-chart: 6.5, CI [6.1, 6.8]cat("Mean efficacy chart: ", format_ci(mean_eff_graph), "\n")## Mean efficacy chart: 6.3, CI [5.9, 6.6]cat("Difference: ", format_ci(mean_eff_diff), "\n")## Difference: -0.24, CI [-0.72, 0.24]# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
ttest <- t.test(eff_graph, eff_nograph)
mean_eff_diff_normal <- c(mean(eff_graph) - mean(eff_nograph), ttest$conf.int[1],
ttest$conf.int[2])
cat("(using a t-test) ", format_ci(mean_eff_diff_normal), "p =", format_number(ttest$p.value),
"\n")## (using a t-test) -0.24, CI [-0.73, 0.26] p = 0.34# Display distributions of efficacy judgments and confidence intervals for
# estimates
## compile CIs and BwS results into a dataframe for plotting
eff_df <- NULL
eff_df <- add.ci.to.df(mean_eff_nograph, "no chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_nochart, bws_nochart, bws_nochart), "no chart",
"BwS", eff_df)
eff_df <- add.ci.to.df(mean_eff_graph, "chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_chart, bws_chart, bws_chart), "chart", "BwS", eff_df)
eff_diff_df <- NULL
eff_diff_df <- add.ci.to.df(mean_eff_diff, " ", "ours")
eff_diff_df <- add.ci.to.df(ci_from_p(bws_chart - bws_nochart, bws_p), " ",
"BwS", eff_diff_df)
if (variable_name == "Belief in efficacy") {
# plotting for experiments 2-4
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp24,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "believe_in_efficacy_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
} else {
# plotting for experiment 1
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.1.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp1,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "perceived_effectiveness_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
}
##### CIs for binary belief in efficacy
# Column is present in experiment 1 only, as subsequent experiments didn't
# include that question.
if ("reduces_sickness" %in% colnames(data)) {
mean_ill_nograph <- propCI(sum(ill_nograph == 1), length(ill_nograph))
mean_ill_graph <- propCI(sum(ill_graph == 1), length(ill_graph))
mean_ill_diff <- diffpropCI(sum(ill_graph == 1), length(ill_graph), sum(ill_nograph ==
1), length(ill_nograph))
print.title("Belief in efficacy (binary)")
cat("Proportion of 'yes' no-chart: ", format_ci(mean_ill_nograph, percent = TRUE),
"\n")
cat("Proportion of 'yes' chart: ", format_ci(mean_ill_graph, percent = TRUE),
"\n")
cat("Difference: ", format_ci(mean_ill_diff, percent = TRUE),
"\n")
# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
yes <- c(sum(ill_graph == 1), sum(ill_nograph == 1))
all <- c(length(ill_graph), length(ill_nograph))
proptest <- prop.test(yes, all, correct = F) # disable continuity correction to get the same p-value
mean_ill_diff_2 <- c(yes[1]/all[1] - yes[2]/all[2], proptest$conf.int[1],
proptest$conf.int[2])
cat("(using an X-squared test) ", format_ci(mean_ill_diff_2, percent = TRUE),
"p =", format_number(proptest$p.value), "\n")
# Plot our results in comparison to the BwS results
# Tal and Wanksink's results
bws.cis.no_graph <- propCI(21, 31)
bws.cis.graph <- propCI(28, 29)
bws.cis.diff <- diffpropCI(28, 29, 21, 31)
# construct the dataframes for plotting
efficiency.df <- NULL
efficiency.df <- add.ci.to.df(mean_ill_nograph, "no chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(mean_ill_graph, "chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.no_graph, "no chart", "BwS", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.graph, "chart", "BwS", efficiency.df)
efficiency.diff.df <- NULL
efficiency.diff.df <- add.ci.to.df(mean_ill_diff, " ", "ours", efficiency.diff.df)
efficiency.diff.df <- add.ci.to.df(bws.cis.diff, " ", "BwS", efficiency.diff.df)
# plot
efficiency.ciplot <- efficacy.belief.ciplot(efficiency.df, efficiency.diff.df)
efficiency.plot <- drawHorizontalEfficiency(efficiency.ciplot, "Belief in efficacy",
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "belief_in_efficacy_plot.pdf"),
efficiency.plot, base_aspect_ratio = 4, base_height = 1.5)
if (print_plots)
print(efficiency.plot)
}Comprehension question
#### CIs for errors at the comprehension question
# Define error
psick <- drug/control
logodds <- function(p) {
log(p/(1 - p))
}
logodds.psick <- logodds(psick)
error <- function(n) {
n[n == 0] <- 0.1
n[n == 20] <- 19.9
abs(logodds(n/20) - logodds.psick)
}
# Compute errors
data$error <- error(data$comprehension)
err_nograph <- error(num_nograph)
err_graph <- error(num_graph)
# Use geometric mean to reduce the influence of outliers and increase the
# weight of close-to-exact answers
mean_err_nograph <- geomMeanCI.bootstrap(err_nograph)
mean_err_graph <- geomMeanCI.bootstrap(err_graph)
mean_err_diff <- ratioGeomMeanCI.bootstrap(err_graph, err_nograph)
print.title("Comprehension error")##
## ###################################
## # Comprehension error
## ###################################cat("Geometric mean error no-chart: ", format_ci(mean_err_nograph), "\n")## Geometric mean error no-chart: 0.83, CI [0.62, 1.06]cat("Geometric mean error chart: ", format_ci(mean_err_graph), "\n")## Geometric mean error chart: 0.75, CI [0.56, 0.97]cat("Ratio: ", format_ci(mean_err_diff), "\n")## Ratio: 0.91, CI [0.63, 1.35]# Plot raw responses and confidence intervals
comp.hist.no_chart <- histo(num_nograph, 20, "no chart", true_answer, 1)
comp.hist.chart <- histo(num_graph, 20, "chart", true_answer, 1)
# construct the dataframes for plotting
comprehension.df <- NULL
comprehension.df <- add.ci.to.df(mean_err_nograph, "no chart", "ours", comprehension.df)
comprehension.df <- add.ci.to.df(mean_err_graph, "chart", "ours", comprehension.df)
comprehension.diff.df <- NULL
comprehension.diff.df <- add.ci.to.df(mean_err_diff, " ", "ours", comprehension.diff.df)
# plot
comp.cis <- comprehension.ciplot(comprehension.df, comprehension.diff.df)
comp.plot <- drawVerticalComp(comp.hist.no_chart, comp.hist.chart, comp.cis,
"Comprehension error", "Error ratio")
if (save_plots) save_plot(paste(sep = "", figure_path, "comprehension_plot.pdf"),
comp.plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots) print(comp.plot)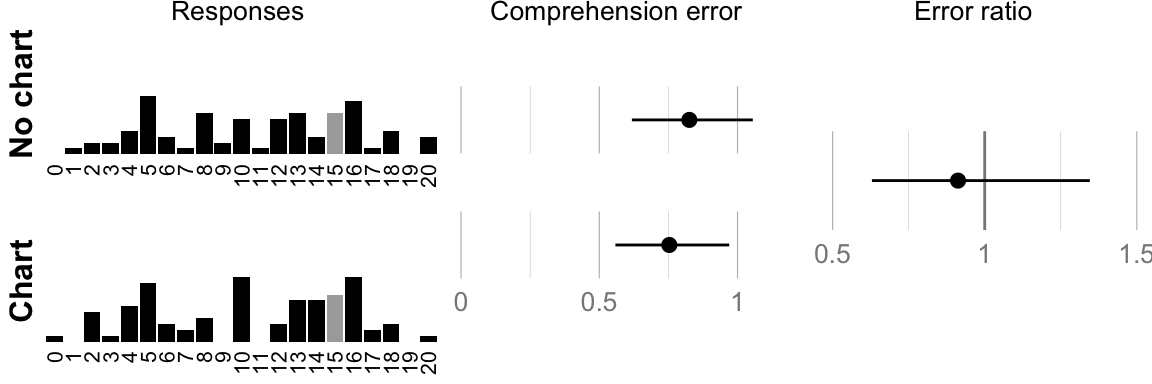
Experiment 3
# Clean up memory
rm(list = ls())
# Experiment filenames
results_file <- "exp3/results"
crowflower_file <- "exp3/crowdflower_data"
invalid_answers_file <- "exp3/invalid_answers"
figure_path <- "plots/exp3-"
# Stimuli values (used to compute comprehension errors)
control <- 83 # % sick people without the drug
drug <- 63 # % sick people with the drug
# compute the true answer given the data
true_answer <- 63/83 * 20
# Values reported by Tal and Wanksink to add to our plots as a comparison
bws_nochart <- 4.66
bws_chart <- 5.75
bws_p <- 0.0066Planned Analysis
library("plyr")
library("dplyr")
source("CI.helpers.R")
source("misc.helpers.R")
source("plotting functions.R")
# indicated whether we want ggplot to save the generated plots
save_plots <- FALSE
# print plots to screen
print_plots <- TRUELoad data
# Load experiment data from csv file
if (exists("results_file")) {
data <- read.csv(paste("data/", results_file, ".csv", sep = ""), sep = ",")
}
# Add crowdflower metadata (if exists)
if (exists("crowflower_file")) {
crowdsourcedata <- read.csv(paste("data/", crowflower_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$completion_code <- as.character(crowdsourcedata$completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "completion_code"))
# Load country codes
country_file <- "countries"
countrydata <- read.csv(paste("data/", country_file, ".csv", sep = ""),
sep = ",")
}
# Add mturk metadata (if exists)
if (exists("mturk_file")) {
crowdsourcedata <- read.csv(paste("data/", mturk_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$Answer.completion_code <- as.character(crowdsourcedata$Answer.completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "Answer.completion_code"))
}Invalid jobs
# Utility function for printing percentages
percent <- function(x, n) {
if (x/n < 0.01)
r <- 1 else r <- 0
paste(" (", format_number(100 * x/n, r), "%)", sep = "")
}
print.title("Invalid jobs")##
## ###################################
## # Invalid jobs
## ###################################data_invalid <- read.csv(paste("data/", invalid_answers_file, ".csv", sep = ""),
sep = ",")
n_valid <- nrow(data)
n_invalid <- nrow(data_invalid)
n <- n_valid + n_invalid
cat("Valid jobs: ", n_valid, "\n")## Valid jobs: 176cat("Invalid jobs: ", n_invalid, "\n\n")## Invalid jobs: 96#
n_failed_attention <- nrow(data_invalid[data_invalid$test_question != "common_cold",
])
n_abnormal_time <- nrow(data_invalid[data_invalid$time_taken < 30 | data_invalid$time_taken >
15 * 60, ])
n_previous_study <- nrow(data_invalid[data_invalid$previous_study == "yes",
])
cat("Failed attention check: ", n_failed_attention, percent(n_failed_attention,
n), "\n", sep = "")## Failed attention check: 30 (10%)cat("Abnormal completion time: ", n_abnormal_time, percent(n_abnormal_time,
n), "\n", sep = "")## Abnormal completion time: 4 (1%)cat("Did study before: ", n_previous_study, percent(n_previous_study,
n), "\n", sep = "")## Did study before: 12 (4%)Sample size, completion time and demographics
print.title("Participants")##
## ###################################
## # Participants
## #################################### Sample size
N <- nrow(data)
cat("Total sample size: ", N, "\n")## Total sample size: 176cat("n =", sum(data$condition == "no_graph"), "for the no-chart condition\n")## n = 88 for the no-chart conditioncat("n =", sum(data$condition == "graph"), "for the chart condition\n\n")## n = 88 for the chart condition# Job completion time
mt <- median(data$time_taken)
cat("Median job completion time:", format_number(mt/60, 2), "minutes.\n\n")## Median job completion time: 2.4 minutes.# Countries
if (exists("crowflower_file")) {
data$country <- sapply(data$X_country, function(code) countrydata$name[match(code,
countrydata$alpha.3)])
data$subregion <- sapply(data$X_country, function(code) countrydata$sub.region[match(code,
countrydata$alpha.3)])
data$continent <- sapply(data$X_country, function(code) countrydata$region[match(code,
countrydata$alpha.3)])
cat("Number of different countries:", length(unique(data$country)), "\n")
print.percents(data$continent)
}## Number of different countries: 40
## n %
## 1 Europe 89 51
## 2 Americas 63 36
## 3 Africa 10 6
## 4 Asia 10 6
## 5 Oceania 2 1# Gender distribution
# Column is present in all but experiment 3
if ("gender" %in% colnames(data)) {
cat("\nGender:\n")
print.percents(data$gender)
}
# Age
# Column is present in all but experiment 3
if ("age" %in% colnames(data)) {
cat("\nMean age:", round(mean(data$age)), "\n")
}
# Education
# Column is present in all but experiment 3
if ("education" %in% colnames(data)) {
cat("\nEducation:\n")
print.percents(data$education)
}Analysis of dependent variables
##### Retrieve data
# Extract judgments of drug efficacy (values between 1 and 9). Experiment 1
# and experiments 2-4 use different question wordings, hence we look at two
# possible columns.
# Column is present in experiment 1, which replicates the question from BwS
# experiment 1
if ("effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$effectiveness
eff_graph <- data[data$condition == "graph", ]$effectiveness
variable_name <- "Perceived effectiveness"
}
# Column is present in experiments 2-4, which replicate the question from
# BwS experiment 2
if ("believe_in_effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$believe_in_effectiveness
eff_graph <- data[data$condition == "graph", ]$believe_in_effectiveness
variable_name <- "Belief in efficacy"
}
# Extract belief in drug from the second question (yes = not effective, no =
# effective).
# Column is present in experiment 1 only.
if ("reduces_sickness" %in% colnames(data)) {
yesno_to_01 <- function(x) {
if (x == "yes")
1 else if (x == "no")
0 else NA
}
ill_nograph <- data[data$condition == "no_graph", ]$reduces_sickness
ill_graph <- data[data$condition == "graph", ]$reduces_sickness
ill_nograph <- sapply(ill_nograph, yesno_to_01)
ill_graph <- sapply(ill_graph, yesno_to_01)
}
# Extract answers to comprehension question
num_nograph <- data[data$condition == "no_graph", ]$comprehension
num_graph <- data[data$condition == "graph", ]$comprehensionBelief in efficacy
##### CIs for judgment of efficacy on a 1--9 scale
mean_eff_nograph <- meanCI.bootstrap(eff_nograph)
mean_eff_graph <- meanCI.bootstrap(eff_graph)
mean_eff_diff <- diffMeanCI.bootstrap(eff_graph, eff_nograph)
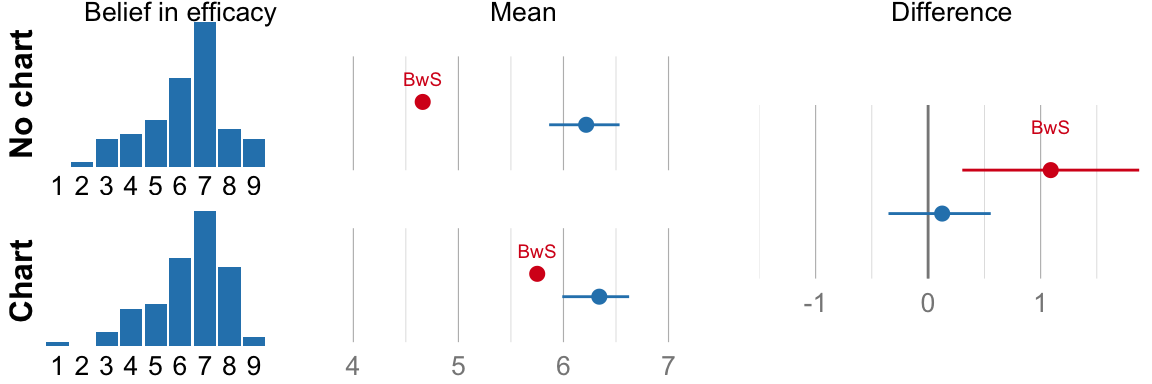
print.title(variable_name)##
## ###################################
## # Belief in efficacy
## ###################################cat("Mean efficacy no-chart: ", format_ci(mean_eff_nograph), "\n")## Mean efficacy no-chart: 6.2, CI [5.9, 6.5]cat("Mean efficacy chart: ", format_ci(mean_eff_graph), "\n")## Mean efficacy chart: 6.3, CI [6.0, 6.6]cat("Difference: ", format_ci(mean_eff_diff), "\n")## Difference: 0.12, CI [-0.35, 0.56]# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
ttest <- t.test(eff_graph, eff_nograph)
mean_eff_diff_normal <- c(mean(eff_graph) - mean(eff_nograph), ttest$conf.int[1],
ttest$conf.int[2])
cat("(using a t-test) ", format_ci(mean_eff_diff_normal), "p =", format_number(ttest$p.value),
"\n")## (using a t-test) 0.12, CI [-0.34, 0.59] p = 0.59# Display distributions of efficacy judgments and confidence intervals for
# estimates
## compile CIs and BwS results into a dataframe for plotting
eff_df <- NULL
eff_df <- add.ci.to.df(mean_eff_nograph, "no chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_nochart, bws_nochart, bws_nochart), "no chart",
"BwS", eff_df)
eff_df <- add.ci.to.df(mean_eff_graph, "chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_chart, bws_chart, bws_chart), "chart", "BwS", eff_df)
eff_diff_df <- NULL
eff_diff_df <- add.ci.to.df(mean_eff_diff, " ", "ours")
eff_diff_df <- add.ci.to.df(ci_from_p(bws_chart - bws_nochart, bws_p), " ",
"BwS", eff_diff_df)
if (variable_name == "Belief in efficacy") {
# plotting for experiments 2-4
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp24,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "believe_in_efficacy_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
} else {
# plotting for experiment 1
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.1.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp1,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "perceived_effectiveness_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
}
##### CIs for binary belief in efficacy
# Column is present in experiment 1 only, as subsequent experiments didn't
# include that question.
if ("reduces_sickness" %in% colnames(data)) {
mean_ill_nograph <- propCI(sum(ill_nograph == 1), length(ill_nograph))
mean_ill_graph <- propCI(sum(ill_graph == 1), length(ill_graph))
mean_ill_diff <- diffpropCI(sum(ill_graph == 1), length(ill_graph), sum(ill_nograph ==
1), length(ill_nograph))
print.title("Belief in efficacy (binary)")
cat("Proportion of 'yes' no-chart: ", format_ci(mean_ill_nograph, percent = TRUE),
"\n")
cat("Proportion of 'yes' chart: ", format_ci(mean_ill_graph, percent = TRUE),
"\n")
cat("Difference: ", format_ci(mean_ill_diff, percent = TRUE),
"\n")
# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
yes <- c(sum(ill_graph == 1), sum(ill_nograph == 1))
all <- c(length(ill_graph), length(ill_nograph))
proptest <- prop.test(yes, all, correct = F) # disable continuity correction to get the same p-value
mean_ill_diff_2 <- c(yes[1]/all[1] - yes[2]/all[2], proptest$conf.int[1],
proptest$conf.int[2])
cat("(using an X-squared test) ", format_ci(mean_ill_diff_2, percent = TRUE),
"p =", format_number(proptest$p.value), "\n")
# Plot our results in comparison to the BwS results
# Tal and Wanksink's results
bws.cis.no_graph <- propCI(21, 31)
bws.cis.graph <- propCI(28, 29)
bws.cis.diff <- diffpropCI(28, 29, 21, 31)
# construct the dataframes for plotting
efficiency.df <- NULL
efficiency.df <- add.ci.to.df(mean_ill_nograph, "no chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(mean_ill_graph, "chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.no_graph, "no chart", "BwS", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.graph, "chart", "BwS", efficiency.df)
efficiency.diff.df <- NULL
efficiency.diff.df <- add.ci.to.df(mean_ill_diff, " ", "ours", efficiency.diff.df)
efficiency.diff.df <- add.ci.to.df(bws.cis.diff, " ", "BwS", efficiency.diff.df)
# plot
efficiency.ciplot <- efficacy.belief.ciplot(efficiency.df, efficiency.diff.df)
efficiency.plot <- drawHorizontalEfficiency(efficiency.ciplot, "Belief in efficacy",
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "belief_in_efficacy_plot.pdf"),
efficiency.plot, base_aspect_ratio = 4, base_height = 1.5)
if (print_plots)
print(efficiency.plot)
}Comprehension question
################################################################################ Comprehension question Display distributions of comprehension test
################################################################################ responses
# par(mfrow=c(1,2)) myhist('Number sick - no chart', num_nograph, 0, 20, 15)
# myhist('Number sick - chart', num_graph, 0, 20, 15)
#### CIs for errors at the comprehension question
# Define error
psick <- drug/control
logodds <- function(p) {
log(p/(1 - p))
}
logodds.psick <- logodds(psick)
error <- function(n) {
n[n == 0] <- 0.1
n[n == 20] <- 19.9
abs(logodds(n/20) - logodds.psick)
}
# Compute errors
data$error <- error(data$comprehension)
err_nograph <- error(num_nograph)
err_graph <- error(num_graph)
# Use geometric mean to reduce the influence of outliers and increase the
# weight of close-to-exact answers
mean_err_nograph <- geomMeanCI.bootstrap(err_nograph)
mean_err_graph <- geomMeanCI.bootstrap(err_graph)
mean_err_diff <- ratioGeomMeanCI.bootstrap(err_graph, err_nograph)
print.title("Comprehension error")##
## ###################################
## # Comprehension error
## ###################################cat("Geometric mean error no-chart: ", format_ci(mean_err_nograph), "\n")## Geometric mean error no-chart: 0.76, CI [0.61, 0.94]cat("Geometric mean error chart: ", format_ci(mean_err_graph), "\n")## Geometric mean error chart: 0.59, CI [0.45, 0.77]cat("Ratio: ", format_ci(mean_err_diff), "\n")## Ratio: 0.78, CI [0.55, 1.08]# Plot raw responses and confidence intervals
comp.hist.no_chart <- histo(num_nograph, 20, "no chart", true_answer, 1)
comp.hist.chart <- histo(num_graph, 20, "chart", true_answer, 1)
# construct the dataframes for plotting
comprehension.df <- NULL
comprehension.df <- add.ci.to.df(mean_err_nograph, "no chart", "ours", comprehension.df)
comprehension.df <- add.ci.to.df(mean_err_graph, "chart", "ours", comprehension.df)
comprehension.diff.df <- NULL
comprehension.diff.df <- add.ci.to.df(mean_err_diff, " ", "ours", comprehension.diff.df)
# plot
comp.cis <- comprehension.ciplot(comprehension.df, comprehension.diff.df)
comp.plot <- drawVerticalComp(comp.hist.no_chart, comp.hist.chart, comp.cis,
"Comprehension error", "Error ratio")
if (save_plots) save_plot(paste(sep = "", figure_path, "comprehension_plot.pdf"),
comp.plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots) print(comp.plot)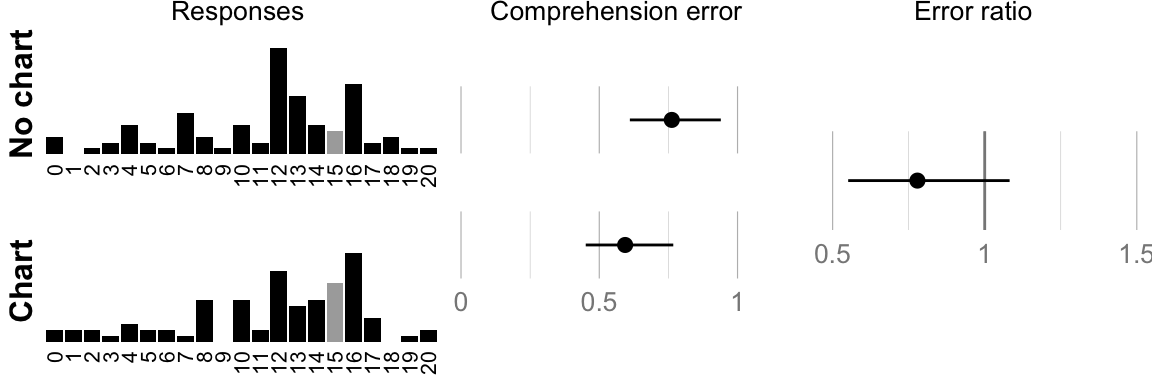
Additional Analysis
Bias in responses
cat("Mean response no-chart: ", format_ci(meanCI.bootstrap(num_nograph), sigdigits = 3),
"\n")## Mean response no-chart: 11.3, CI [10.2, 12.2]cat("Mean response chart: ", format_ci(meanCI.bootstrap(num_graph), sigdigits = 3),
"\n")## Mean response chart: 11.8, CI [10.7, 12.7]cat("Mean difference: ", format_ci(diffMeanCI.bootstrap(num_graph, num_nograph)),
"\n")## Mean difference: 0.48, CI [-0.91, 1.85]Experiment 4
# Clean up memory
rm(list = ls())
# Experiment filenames
results_file <- "exp4/results"
mturk_file <- "exp4/mturk"
invalid_answers_file <- "exp4/invalid_answers"
figure_path <- "plots/exp4-"
# Stimuli values (used to compute comprehension errors)
control <- 83 # % sick people without the drug
drug <- 63 # % sick people with the drug
# compute the true answer given the data
true_answer <- 63/83 * 20
# Values reported by Tal and Wanksink to add to our plots as a comparison
bws_nochart <- 4.66
bws_chart <- 5.75
bws_p <- 0.0066Planned Analysis
library("plyr")
library("dplyr")
source("CI.helpers.R")
source("misc.helpers.R")
source("plotting functions.R")
# indicated whether we want ggplot to save the generated plots
save_plots <- FALSE
# print plots to screen
print_plots <- TRUELoad data
# Load experiment data from csv file
if (exists("results_file")) {
data <- read.csv(paste("data/", results_file, ".csv", sep = ""), sep = ",")
}
# Add crowdflower metadata (if exists)
if (exists("crowflower_file")) {
crowdsourcedata <- read.csv(paste("data/", crowflower_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$completion_code <- as.character(crowdsourcedata$completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "completion_code"))
# Load country codes
country_file <- "countries"
countrydata <- read.csv(paste("data/", country_file, ".csv", sep = ""),
sep = ",")
}
# Add mturk metadata (if exists)
if (exists("mturk_file")) {
crowdsourcedata <- read.csv(paste("data/", mturk_file, ".csv", sep = ""),
sep = ",")
data$code <- as.character(data$code)
crowdsourcedata$Answer.completion_code <- as.character(crowdsourcedata$Answer.completion_code)
data <- left_join(data, crowdsourcedata, by = c(code = "Answer.completion_code"))
}Invalid jobs
# Utility function for printing percentages
percent <- function(x, n) {
if (x/n < 0.01)
r <- 1 else r <- 0
paste(" (", format_number(100 * x/n, r), "%)", sep = "")
}
data_invalid <- read.csv(paste("data/", invalid_answers_file, ".csv", sep = ""),
sep = ",")
n_valid <- nrow(data)
n_invalid <- nrow(data_invalid)
n <- n_valid + n_invalid
cat("Valid jobs: ", n_valid, "\n")## Valid jobs: 160cat("Invalid jobs: ", n_invalid, "\n\n")## Invalid jobs: 29#
n_failed_attention <- nrow(data_invalid[data_invalid$test_question != "common_cold",
])
n_abnormal_time <- nrow(data_invalid[data_invalid$time_taken < 30 | data_invalid$time_taken >
15 * 60, ])
n_previous_study <- nrow(data_invalid[data_invalid$previous_study == "yes",
])
cat("Failed attention check: ", n_failed_attention, percent(n_failed_attention,
n), "\n", sep = "")## Failed attention check: 6 (3%)cat("Abnormal completion time: ", n_abnormal_time, percent(n_abnormal_time,
n), "\n", sep = "")## Abnormal completion time: 0 (0%)cat("Did study before: ", n_previous_study, percent(n_previous_study,
n), "\n", sep = "")## Did study before: 0 (0%)Sample size, completion time and demographics
print.title("Participants")##
## ###################################
## # Participants
## #################################### Sample size
N <- nrow(data)
cat("Total sample size: ", N, "\n")## Total sample size: 160cat("n =", sum(data$condition == "no_graph"), "for the no-chart condition\n")## n = 80 for the no-chart conditioncat("n =", sum(data$condition == "graph"), "for the chart condition\n\n")## n = 80 for the chart condition# Job completion time
mt <- median(data$time_taken)
cat("Median job completion time:", format_number(mt/60, 2), "minutes.\n\n")## Median job completion time: 1.7 minutes.# Countries
if (exists("crowflower_file")) {
data$country <- sapply(data$X_country, function(code) countrydata$name[match(code,
countrydata$alpha.3)])
data$subregion <- sapply(data$X_country, function(code) countrydata$sub.region[match(code,
countrydata$alpha.3)])
data$continent <- sapply(data$X_country, function(code) countrydata$region[match(code,
countrydata$alpha.3)])
cat("Number of different countries:", length(unique(data$country)), "\n")
print.percents(data$continent)
}
# Gender distribution
# Column is present in all but experiment 3
if ("gender" %in% colnames(data)) {
cat("\nGender:\n")
print.percents(data$gender)
}##
## Gender:
## n %
## 1 male 89 56
## 2 female 70 44
## 3 no_answer 1 1# Age
# Column is present in all but experiment 3
if ("age" %in% colnames(data)) {
cat("\nMean age:", round(mean(data$age)), "\n")
}##
## Mean age: 37# Education
# Column is present in all but experiment 3
if ("education" %in% colnames(data)) {
cat("\nEducation:\n")
print.percents(data$education)
}##
## Education:
## n %
## 1 college_or_more 77 48
## 2 some_college 60 38
## 3 high-school 23 14Analysis of dependent variables
##### Retrieve data
# Extract judgments of drug efficacy (values between 1 and 9). Experiment 1
# and experiments 2-4 use different question wordings, hence we look at two
# possible columns.
# Column is present in experiment 1, which replicates the question from BwS
# experiment 1
if ("effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$effectiveness
eff_graph <- data[data$condition == "graph", ]$effectiveness
variable_name <- "Perceived effectiveness"
}
# Column is present in experiments 2-4, which replicate the question from
# BwS experiment 2
if ("believe_in_effectiveness" %in% colnames(data)) {
eff_nograph <- data[data$condition == "no_graph", ]$believe_in_effectiveness
eff_graph <- data[data$condition == "graph", ]$believe_in_effectiveness
variable_name <- "Belief in efficacy"
}
# Extract belief in drug from the second question (yes = not effective, no =
# effective).
# Column is present in experiment 1 only.
if ("reduces_sickness" %in% colnames(data)) {
yesno_to_01 <- function(x) {
if (x == "yes")
1 else if (x == "no")
0 else NA
}
ill_nograph <- data[data$condition == "no_graph", ]$reduces_sickness
ill_graph <- data[data$condition == "graph", ]$reduces_sickness
ill_nograph <- sapply(ill_nograph, yesno_to_01)
ill_graph <- sapply(ill_graph, yesno_to_01)
}
# Extract answers to comprehension question
num_nograph <- data[data$condition == "no_graph", ]$comprehension
num_graph <- data[data$condition == "graph", ]$comprehensionBelief in Efficacy
##### CIs for judgment of efficacy on a 1--9 scale
mean_eff_nograph <- meanCI.bootstrap(eff_nograph)
mean_eff_graph <- meanCI.bootstrap(eff_graph)
mean_eff_diff <- diffMeanCI.bootstrap(eff_graph, eff_nograph)
print.title(variable_name)##
## ###################################
## # Belief in efficacy
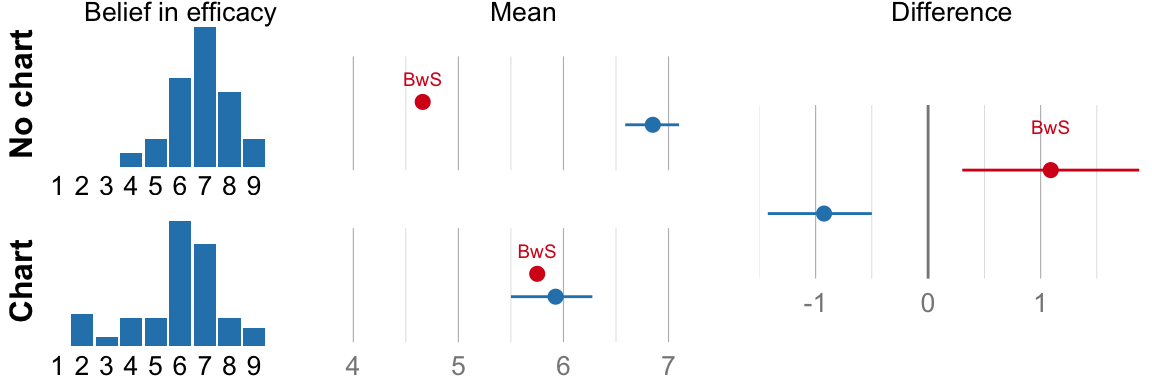
## ###################################cat("Mean efficacy no-chart: ", format_ci(mean_eff_nograph), "\n")## Mean efficacy no-chart: 6.8, CI [6.6, 7.1]cat("Mean efficacy chart: ", format_ci(mean_eff_graph), "\n")## Mean efficacy chart: 5.9, CI [5.5, 6.3]cat("Difference: ", format_ci(mean_eff_diff), "\n")## Difference: -0.92, CI [-1.42, -0.50]# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
ttest <- t.test(eff_graph, eff_nograph)
mean_eff_diff_normal <- c(mean(eff_graph) - mean(eff_nograph), ttest$conf.int[1],
ttest$conf.int[2])
cat("(using a t-test) ", format_ci(mean_eff_diff_normal), "p =", format_number(ttest$p.value),
"\n")## (using a t-test) -0.92, CI [-1.39, -0.46] p = 0.00013# Display distributions of efficacy judgments and confidence intervals for
# estimates
## compile CIs and BwS results into a dataframe for plotting
eff_df <- NULL
eff_df <- add.ci.to.df(mean_eff_nograph, "no chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_nochart, bws_nochart, bws_nochart), "no chart",
"BwS", eff_df)
eff_df <- add.ci.to.df(mean_eff_graph, "chart", "ours", eff_df)
eff_df <- add.ci.to.df(c(bws_chart, bws_chart, bws_chart), "chart", "BwS", eff_df)
eff_diff_df <- NULL
eff_diff_df <- add.ci.to.df(mean_eff_diff, " ", "ours")
eff_diff_df <- add.ci.to.df(ci_from_p(bws_chart - bws_nochart, bws_p), " ",
"BwS", eff_diff_df)
if (variable_name == "Belief in efficacy") {
# plotting for experiments 2-4
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp24,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "believe_in_efficacy_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
} else {
# plotting for experiment 1
eff.hist.no_chart <- histo(eff_nograph, 9, "no chart", 0, 1)
eff.hist.chart <- histo(eff_graph, 9, "chart", 0, 1)
eff.cis <- perceived.effectiveness.1.ciplot(eff_df, eff_diff_df)
plot <- drawVertical(eff.hist.no_chart, eff.hist.chart, eff.cis, name.eff.exp1,
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "perceived_effectiveness_plot.pdf"),
plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots)
print(plot)
}
##### CIs for binary belief in efficacy
# Column is present in experiment 1 only, as subsequent experiments didn't
# include that question.
if ("reduces_sickness" %in% colnames(data)) {
mean_ill_nograph <- propCI(sum(ill_nograph == 1), length(ill_nograph))
mean_ill_graph <- propCI(sum(ill_graph == 1), length(ill_graph))
mean_ill_diff <- diffpropCI(sum(ill_graph == 1), length(ill_graph), sum(ill_nograph ==
1), length(ill_nograph))
print.title("Belief in efficacy (binary)")
cat("Proportion of 'yes' no-chart: ", format_ci(mean_ill_nograph, percent = TRUE),
"\n")
cat("Proportion of 'yes' chart: ", format_ci(mean_ill_graph, percent = TRUE),
"\n")
cat("Difference: ", format_ci(mean_ill_diff, percent = TRUE),
"\n")
# NHST version for comparison (used in the original paper, but not part of
# this planned analysis)
yes <- c(sum(ill_graph == 1), sum(ill_nograph == 1))
all <- c(length(ill_graph), length(ill_nograph))
proptest <- prop.test(yes, all, correct = F) # disable continuity correction to get the same p-value
mean_ill_diff_2 <- c(yes[1]/all[1] - yes[2]/all[2], proptest$conf.int[1],
proptest$conf.int[2])
cat("(using an X-squared test) ", format_ci(mean_ill_diff_2, percent = TRUE),
"p =", format_number(proptest$p.value), "\n")
# Plot our results in comparison to the BwS results
# Tal and Wanksink's results
bws.cis.no_graph <- propCI(21, 31)
bws.cis.graph <- propCI(28, 29)
bws.cis.diff <- diffpropCI(28, 29, 21, 31)
# construct the dataframes for plotting
efficiency.df <- NULL
efficiency.df <- add.ci.to.df(mean_ill_nograph, "no chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(mean_ill_graph, "chart", "ours", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.no_graph, "no chart", "BwS", efficiency.df)
efficiency.df <- add.ci.to.df(bws.cis.graph, "chart", "BwS", efficiency.df)
efficiency.diff.df <- NULL
efficiency.diff.df <- add.ci.to.df(mean_ill_diff, " ", "ours", efficiency.diff.df)
efficiency.diff.df <- add.ci.to.df(bws.cis.diff, " ", "BwS", efficiency.diff.df)
# plot
efficiency.ciplot <- efficacy.belief.ciplot(efficiency.df, efficiency.diff.df)
efficiency.plot <- drawHorizontalEfficiency(efficiency.ciplot, "Belief in efficacy",
"Difference")
if (save_plots)
save_plot(paste(sep = "", figure_path, "belief_in_efficacy_plot.pdf"),
efficiency.plot, base_aspect_ratio = 4, base_height = 1.5)
if (print_plots)
print(efficiency.plot)
}Comprehension question
# Display distributions of comprehension test responses
# par(mfrow=c(1,2)) myhist('Number sick - no chart', num_nograph, 0, 20, 15)
# myhist('Number sick - chart', num_graph, 0, 20, 15)
#### CIs for errors at the comprehension question
# Define error
psick <- drug/control
logodds <- function(p) {
log(p/(1 - p))
}
logodds.psick <- logodds(psick)
error <- function(n) {
n[n == 0] <- 0.1
n[n == 20] <- 19.9
abs(logodds(n/20) - logodds.psick)
}
# Compute errors
data$error <- error(data$comprehension)
err_nograph <- error(num_nograph)
err_graph <- error(num_graph)
# Use geometric mean to reduce the influence of outliers and increase the
# weight of close-to-exact answers
mean_err_nograph <- geomMeanCI.bootstrap(err_nograph)
mean_err_graph <- geomMeanCI.bootstrap(err_graph)
mean_err_diff <- ratioGeomMeanCI.bootstrap(err_graph, err_nograph)
print.title("Comprehension error")##
## ###################################
## # Comprehension error
## ###################################cat("Geometric mean error no-chart: ", format_ci(mean_err_nograph), "\n")## Geometric mean error no-chart: 0.47, CI [0.36, 0.60]cat("Geometric mean error chart: ", format_ci(mean_err_graph), "\n")## Geometric mean error chart: 0.46, CI [0.35, 0.59]cat("Ratio: ", format_ci(mean_err_diff), "\n")## Ratio: 0.97, CI [0.69, 1.42]# Plot raw responses and confidence intervals
comp.hist.no_chart <- histo(num_nograph, 20, "no chart", true_answer, 1)
comp.hist.chart <- histo(num_graph, 20, "chart", true_answer, 1)
# construct the dataframes for plotting
comprehension.df <- NULL
comprehension.df <- add.ci.to.df(mean_err_nograph, "no chart", "ours", comprehension.df)
comprehension.df <- add.ci.to.df(mean_err_graph, "chart", "ours", comprehension.df)
comprehension.diff.df <- NULL
comprehension.diff.df <- add.ci.to.df(mean_err_diff, " ", "ours", comprehension.diff.df)
# plot
comp.cis <- comprehension.ciplot(comprehension.df, comprehension.diff.df)
comp.plot <- drawVerticalComp(comp.hist.no_chart, comp.hist.chart, comp.cis,
"Comprehension error", "Error ratio")
if (save_plots) save_plot(paste(sep = "", figure_path, "comprehension_plot.pdf"),
comp.plot, base_aspect_ratio = 3, base_height = 2)
if (print_plots) print(comp.plot)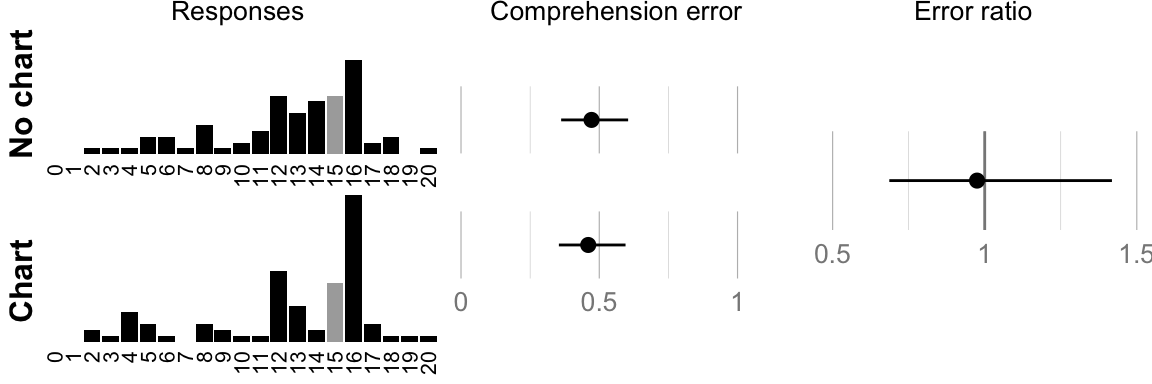
Extra planned analysis
# Remove NAs for belief in science (not sure how this happened)
data_clean <- data[!is.na(data$belief_in_science), ]
# Break down by condition
nograph <- data_clean[data_clean$condition == "no_graph", ]
graph <- data_clean[data_clean$condition == "graph", ]
# Datasets with positively polarized people removed
nograph_nopp <- nograph[nograph$belief_in_medicine < 7, ]
graph_nopp <- graph[graph$belief_in_medicine < 7, ]
# Keep the removed people to plot separately
nograph_removed <- nograph[nograph$belief_in_medicine >= 7, ]
graph_removed <- graph[graph$belief_in_medicine >= 7, ]
# Function for computing and plotting the regression
fit <- function(title, x, xs, y, ys, x_ignored = NULL, y_ignored = NULL) {
coeff_bigger = 1.2
exponent = 0.5
AA = xyTable(x, y)
plot(AA$x, AA$y, main = title, xlab = xs, ylab = ys, xlim = c(1, 9), xaxp = c(1,
9, 8), ylim = c(1, 9), yaxp = c(1, 9, 8), yaxs = "r", type = "n")
fit <- lm(y ~ x)
clip(min(AA$x) - 0.5, max(AA$x) + 0.5, 0, 10)
abline(fit, lty = 1)
clip(0, 10, 0, 10)
confint(fit)
points(AA$x, AA$y, cex = (AA$number)^exponent * coeff_bigger, col = rgb(0,
0, 0, 0.2), pch = 16)
text(AA$x, AA$y, ifelse(AA$number > 1, AA$number, ""), col = rgb(0, 0, 0,
0.5))
if (length(x_ignored) > 0) {
AA = xyTable(x_ignored, y_ignored)
points(AA$x, AA$y, cex = (AA$number)^exponent * coeff_bigger, col = rgb(0,
0, 0, 0.2), pch = 1)
text(AA$x, AA$y, ifelse(AA$number > 1, AA$number, ""), col = rgb(0,
0, 0, 0.3))
}
}
# Interaction between belief in science and chart persuasiveness (Fig 16
# left part)
layout(matrix(c(1, 2, 3, 4), 1, 4, byrow = T), widths = c(2.5, 2.5, 2.5, 2.5),
heights = c(2.5))
fit("No chart", nograph$belief_in_science, "Belief in science", nograph$believe_in_effectiveness,
"Belief in efficacy")
fit("Chart", graph$belief_in_science, "Belief in science", graph$believe_in_effectiveness,
"Belief in efficacy")
# Interaction between polarization and chart persuasiveness (Fig 16 right
# part)
fit("No chart", nograph_nopp$belief_in_medicine, "Polarization", nograph_nopp$believe_in_effectiveness,
"Belief in efficacy", nograph_removed$belief_in_medicine, nograph_removed$believe_in_effectiveness)
fit("Chart", graph_nopp$belief_in_medicine, "Polarization", graph_nopp$believe_in_effectiveness,
"Belief in efficacy", graph_removed$belief_in_medicine, graph_removed$believe_in_effectiveness)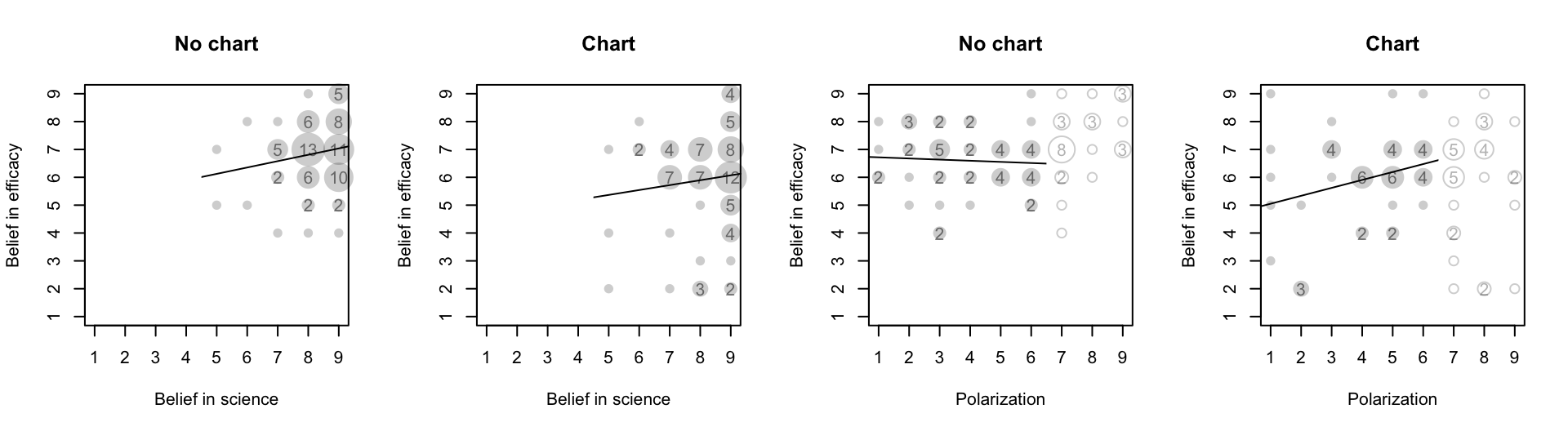
Left group: interaction betweeen belief in science and chart persuasiveness Right group: interaction between polarization and chart persuasiveness
Regression analysis
# Difference between slopes: belief in science and chart persuasiveness
cat("Difference between slopes -- belief in science: ", format_ci(diff.slopes.bootstrap(nograph$belief_in_science,
nograph$believe_in_effectiveness, graph$belief_in_science, graph$believe_in_effectiveness)),
"\n")## Difference between slopes -- belief in science: -0.052, CI [-0.61, 0.37]cat("Difference between slopes -- polarization: ", format_ci(diff.slopes.bootstrap(nograph_nopp$belief_in_medicine,
nograph_nopp$believe_in_effectiveness, graph_nopp$belief_in_medicine, graph_nopp$believe_in_effectiveness)),
"\n")## Warning in norm.inter(t, adj.alpha): extreme order statistics used as
## endpoints## Difference between slopes -- polarization: 0.33, CI [0.31, 0.75]Meta Analysis
# Clean up memory
rm(list = ls())
library("dplyr")
library("plyr")
library("bootES")
source("CI.helpers.R")
source("misc.helpers.R")
# Number of replicates to use in bootstrapping. The higher the more accurate
# the results. Warning: a large R can yield very lengthy computations.
R = 5000
######################################################################## Load data
# Read data from all 4 experiments
data1 <- read.csv(paste("data/exp1/results.csv", sep = ""), sep = ",")
data2 <- read.csv(paste("data/exp2/results.csv", sep = ""), sep = ",")
data3 <- read.csv(paste("data/exp3/results.csv", sep = ""), sep = ",")
data4 <- read.csv(paste("data/exp4/results.csv", sep = ""), sep = ",")
# Compute the total number of observations
N <- nrow(data1) + nrow(data2) + nrow(data3) + nrow(data4)
cat("Total sample size: N =", N, "\n\n")## Total sample size: N = 623######################################################################## Retrieve observations of interest
# Efficacy judgement data for all 4 experiments
eff1_nograph <- data1[data1$condition == "no_graph", ]$effectiveness
eff2_nograph <- data2[data2$condition == "no_graph", ]$believe_in_effectiveness
eff3_nograph <- data3[data3$condition == "no_graph", ]$believe_in_effectiveness
eff4_nograph <- data4[data4$condition == "no_graph", ]$believe_in_effectiveness
eff1_graph <- data1[data1$condition == "graph", ]$effectiveness
eff2_graph <- data2[data2$condition == "graph", ]$believe_in_effectiveness
eff3_graph <- data3[data3$condition == "graph", ]$believe_in_effectiveness
eff4_graph <- data4[data4$condition == "graph", ]$believe_in_effectiveness
# Comprehension error data for all 4 experiments
logodds <- function(p) {
log(p/(1 - p))
}
error <- function(n, control, drug) {
n[n == 0] <- 0.1
n[n == 20] <- 19.9
abs(logodds(n/20) - logodds(drug/control))
}
error1 <- function(n) {
error(n, 87, 47)
} # For exp 1
error2 <- function(n) {
error(n, 83, 63)
} # For exp 2,3,4
err1_nograph <- error1(data1[data1$condition == "no_graph", ]$comprehension)
err2_nograph <- error2(data2[data2$condition == "no_graph", ]$comprehension)
err3_nograph <- error2(data3[data3$condition == "no_graph", ]$comprehension)
err4_nograph <- error2(data4[data4$condition == "no_graph", ]$comprehension)
err1_graph <- error1(data1[data1$condition == "graph", ]$comprehension)
err2_graph <- error2(data2[data2$condition == "graph", ]$comprehension)
err3_graph <- error2(data3[data3$condition == "graph", ]$comprehension)
err4_graph <- error2(data4[data4$condition == "graph", ]$comprehension)
######################################################################## Compute standardized effect sizes and their CI for each experiment
# Convenience function that computes, prints and returns the standardized
# effect size for the mean difference between two independent samples, with
# its 95% BCa bootstrap CI.
printEffectSize <- function(exp_name, data_nograph, data_graph) {
data <- data.frame(obs = c(data_nograph, data_graph), group = c(rep("nograph",
length(data_nograph)), rep("graph", length(data_graph))))
set.seed(0) # make deterministic
b <- bootES(data, R = R, effect.type = "cohens.d", data.col = "obs", group.col = "group",
contrast = c(nograph = 1, graph = -1))
pointEstimate <- b[[1]]
bootci <- boot.ci(b, type = "bca", conf = conf.level)
cat("Effect size for ", exp_name, ": ", format_ci(c(pointEstimate, bootci$bca[4],
bootci$bca[5])), "\n", sep = "")
return(data.frame(estimate = pointEstimate, ci.low = bootci$bca[4], ci.high = bootci$bca[5]))
}
# Note: the exact point and interval estimates obtained here may differ
# slightly from the paper because in the previous version, the random seed
# was not reset before each call to bootES. Use a large value of R to get
# more accurate estimates.
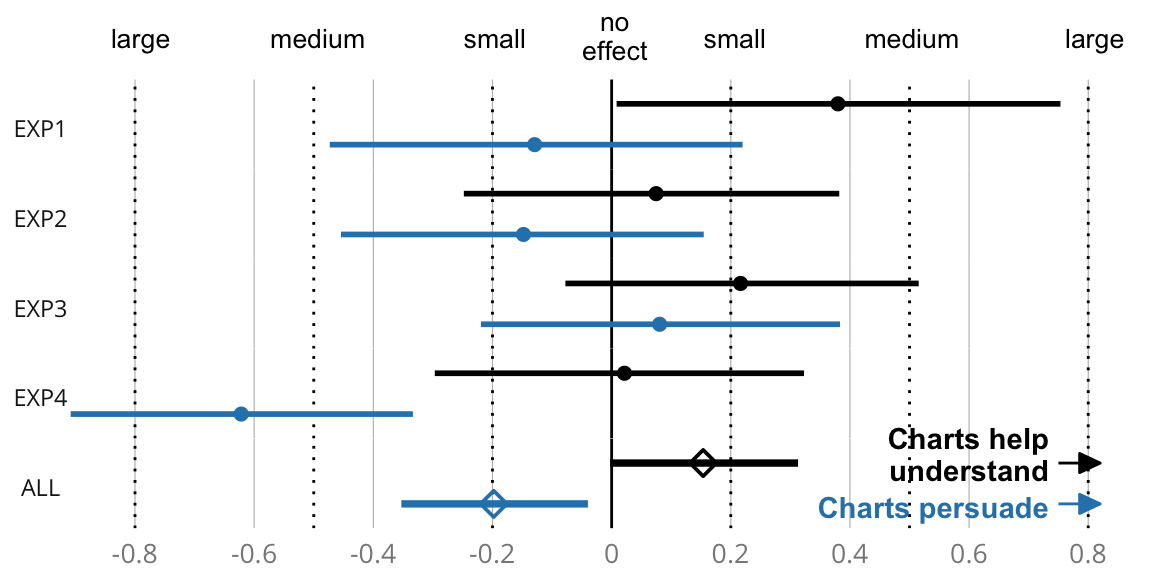
exp1.belief <- printEffectSize("exp 1 belief ", eff1_graph, eff1_nograph)## Effect size for exp 1 belief : -0.13, CI [-0.47, 0.22]exp2.belief <- printEffectSize("exp 2 belief ", eff2_graph, eff2_nograph)## Effect size for exp 2 belief : -0.15, CI [-0.45, 0.15]exp3.belief <- printEffectSize("exp 3 belief ", eff3_graph, eff3_nograph)## Effect size for exp 3 belief : 0.081, CI [-0.22, 0.38]exp4.belief <- printEffectSize("exp 4 belief ", eff4_graph, eff4_nograph)## Effect size for exp 4 belief : -0.62, CI [-0.91, -0.33]cat("\n")exp1.error <- printEffectSize("exp 1 log error ", log(err1_nograph), log(err1_graph))## Effect size for exp 1 log error : 0.38, CI [0.0084, 0.75]exp2.error <- printEffectSize("exp 2 log error ", log(err2_nograph), log(err2_graph))## Effect size for exp 2 log error : 0.075, CI [-0.25, 0.38]exp3.error <- printEffectSize("exp 3 log error ", log(err3_nograph), log(err3_graph))## Effect size for exp 3 log error : 0.22, CI [-0.078, 0.52]exp4.error <- printEffectSize("exp 4 log error ", log(err4_nograph), log(err4_graph))## Effect size for exp 4 log error : 0.022, CI [-0.30, 0.32]cat("\n")######################################################################## Compute the aggregated standardized effect size and its CI
# Custom function that performs a contrast weighted by sample size to
# compute a standardized effect size for all 4 experiments together, and its
# 95% BCa bootstrap CI. ng1: no-graph condition, experiment 1 ng2: no-graph
# condition, experiment 2 ng3: no-graph condition, experiment 3 ng4:
# no-graph condition, experiment 4 g1: graph condition, experiment 1 g2:
# graph condition, experiment 2 g3: graph condition, experiment 3 g4: graph
# condition, experiment 4
printMetaEffectSize <- function(exp, ng1, ng2, ng3, ng4, g1, g2, g3, g4) {
# Create custom data frame to hold all the data
data <- data.frame(obs = c(ng1, ng2, ng3, ng4, g1, g2, g3, g4), group = c(rep("ng1",
length(ng1)), rep("ng2", length(ng2)), rep("ng3", length(ng3)), rep("ng4",
length(ng4)), rep("g1", length(g1)), rep("g2", length(g2)), rep("g3",
length(g3)), rep("g4", length(g4))))
# Set weights for the contrast
n_ng = length(ng1) + length(ng2) + length(ng3) + length(ng4) # total sample size for no-graph
n_g = length(g1) + length(g2) + length(g3) + length(g4) # total sample size for graph
w_ng1 = length(ng1)/n_ng
w_ng2 = length(ng2)/n_ng
w_ng3 = length(ng3)/n_ng
w_ng4 = length(ng4)/n_ng
w_g1 = length(g1)/n_g
w_g2 = length(g2)/n_g
w_g3 = length(g3)/n_g
w_g4 = length(g4)/n_g
# print(w_ng1+w_ng2+w_ng3-w_g1-w_g2-w_g3) # should be zero
set.seed(0) # make deterministic
b <- bootES(data, R = R, effect.type = "cohens.d", data.col = "obs", group.col = "group",
scale.weights = T, contrast = c(ng1 = w_ng1, ng2 = w_ng2, ng3 = w_ng3,
ng4 = w_ng4, g1 = -w_g1, g2 = -w_g2, g3 = -w_g3, g4 = -w_g4))
pointEstimate <- b[[1]]
bootci <- boot.ci(b, type = "bca", conf = conf.level)
cat("Effect size for ", exp, ": ", format_ci(c(pointEstimate, bootci$bca[4],
bootci$bca[5])), "\n", sep = "")
return(data.frame(estimate = pointEstimate, ci.low = bootci$bca[4], ci.high = bootci$bca[5]))
}
# Note: the exact point and interval estimates obtained here may differ
# slightly from the paper because in the previous version, the random seed
# was not reset before each call to bootES. Use a large value of R to get
# more accurate estimates.
meta.belief <- printMetaEffectSize("all belief ", eff1_graph, eff2_graph,
eff3_graph, eff4_graph, eff1_nograph, eff2_nograph, eff3_nograph, eff4_nograph)## Effect size for all belief : -0.20, CI [-0.35, -0.040]cat("\n")meta.error <- printMetaEffectSize("all log error ", log(err1_nograph), log(err2_nograph),
log(err3_nograph), log(err4_nograph), log(err1_graph), log(err2_graph),
log(err3_graph), log(err4_graph))## Effect size for all log error : 0.15, CI [-0.0022, 0.31]library('ggplot2')
library("gridExtra")
library("cowplot")
source("CI.helpers.R")
source("misc.helpers.R")
scale.min <- -0.92
scale.max <- 0.82
effect.size.indicator.color <- "black"
meta.ci.df <- NULL
meta.ci.df <- add.ci.to.df(exp1.belief, "belief", "EXP1", meta.ci.df)
meta.ci.df <- add.ci.to.df(exp1.error, "error", "EXP1", meta.ci.df)
meta.ci.df <- add.ci.to.df(exp2.belief, "belief", "EXP2", meta.ci.df)
meta.ci.df <- add.ci.to.df(exp2.error, "error", "EXP2", meta.ci.df)
meta.ci.df <- add.ci.to.df(exp3.belief, "belief", "EXP3", meta.ci.df)
meta.ci.df <- add.ci.to.df(exp3.error, "error", "EXP3", meta.ci.df)
meta.ci.df <- add.ci.to.df(exp4.belief, "belief", "EXP4", meta.ci.df)
meta.ci.df <- add.ci.to.df(exp4.error, "error", "EXP4", meta.ci.df)
meta.ci.df <- add.ci.to.df(meta.belief, "belief", "ALL", meta.ci.df)
meta.ci.df <- add.ci.to.df(meta.error, "error", "ALL", meta.ci.df)
citheme <- theme_bw(base_family = "Open Sans") +
theme( # remove the vertical grid lines
plot.background = element_blank(),
plot.margin = margin(t = 0, r = 0, b = 0, l = 0, unit = "pt"),
legend.position = "none", #c(0.2,0.88),
legend.title = element_blank(),
legend.margin = margin(t = 0, r = 0, b = 0, l = 0, unit = "pt"),
legend.text = element_text(size=8),
legend.key.size = unit(c(3.5, 3.5),'mm'),
legend.key.width = unit(c(0.3, 0.3),'cm'),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_blank(),
axis.text.x = element_text(size=10, colour="#888888", margin = margin(1,1.5,0,0,"mm")),
panel.spacing = unit(0, "lines"),
panel.border = element_blank(),
axis.line = element_blank(),
axis.ticks = element_blank(),
axis.line.x = element_blank(),
panel.grid.minor.x = element_blank(),
panel.grid.major.x = element_line(size=0.2, color="#BBBBBB"),
panel.grid.major.y = element_blank(),
strip.background = element_blank(),
strip.text.y = element_text(angle = 180, margin = margin(1,-7,0,0,"mm"))
)
condFatten <- function(x,...) {
ifelse(x=="ALL", 1.3, 1)
}
condShape <- function(x,...) {
ifelse(x=="ALL", 5, 16)
}
meta.ciplot <- function(df, scale = T) {
ratioPlot <- ggplot(data= df) +
expand_limits( y=c(scale.min, scale.max)) +
citheme +
geom_hline(yintercept = -0.8, color = effect.size.indicator.color, linetype = "dotted") +
geom_hline(yintercept = -0.5, color = effect.size.indicator.color, linetype = "dotted") +
geom_hline(yintercept = -0.2, color = effect.size.indicator.color, linetype = "dotted") +
geom_hline(yintercept = 0, color = "black") +
geom_hline(yintercept = 0.2, color = effect.size.indicator.color, linetype = "dotted") +
geom_hline(yintercept = 0.5, color = effect.size.indicator.color, linetype = "dotted") +
geom_hline(yintercept = 0.8, color = effect.size.indicator.color, linetype = "dotted") +
coord_flip() +
geom_pointrange(aes(y = estimate, x=reorder(name, order), ymin= ci.low, ymax = ci.high, color = name), size = condFatten(df$study), fatten = 2, shape= condShape(df$study)) +
scale_colour_manual(name = 'belief in blue', values = setNames(c('#2b83ba', "black", "black", "black"),c("belief", "error", "meta", "afake"))) +
geom_path(data=data.frame(name=c("error", "error", "belief", "belief"), estimate = c(0.75, 0.82, 0.75, 0.82), study= c("ALL", "ALL", "ALL", "ALL")), aes(x = name, y = estimate, color=name), arrow=arrow(angle = 25, length = unit(3, "mm"), type="closed")) +
facet_grid(study ~ ., space= "free_x", switch = "y") +
labs(x=NULL, y=NULL) +
theme(axis.text.y = element_blank()) +
scale_y_continuous(breaks= seq(from=-0.8, to=0.8, by= 0.2), labels = seq(from=-0.8, to=0.8, by= 0.2), limits = c(scale.min, scale.max))
if (!scale) {
ratioPlot <- ratioPlot + theme(axis.text.x = element_blank() )
}
return(ratioPlot)
}
metaplot <- meta.ciplot(meta.ci.df)
finalplot <- ggdraw() +
draw_plot(metaplot, 0, 0, 1, 0.86) +
draw_label("large", 0.115, 0.95, vjust=1, hjust = 0.5, size = 10) +
draw_label("medium", 0.27, 0.95, vjust=1, hjust = 0.5, size = 10) +
draw_label("small", 0.425, 0.95, vjust=1, hjust = 0.5, size = 10) +
draw_label("no\neffect", 0.53, 0.98, vjust=1, hjust = 0.5, size = 10) +
draw_label("small", 0.635, 0.95, vjust=1, hjust = 0.5, size = 10) +
draw_label("medium", 0.79, 0.95, vjust=1, hjust = 0.5, size = 10) +
draw_label("large", 0.95, 0.95, vjust=1, hjust = 0.5, size = 10) +
draw_label("Charts help\nunderstand", 0.91, 0.06, vjust = -1, hjust = 1, size = 11, fontface = "bold") +
draw_label("Charts persuade", 0.91, 0.05, vjust = -1, hjust = 1, size = 11, colour = '#2b83ba', fontface = "bold") ## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font database
## Warning in grid.Call(L_textBounds, as.graphicsAnnot(x$label), x$x, x$y, :
## font family 'Open Sans' not found in PostScript font databaseprint(finalplot)